Abstract
Acute Myeloid Leukemia (AML) remains notoriously difficult to treat. AMLs manifest themselves within individual patients with remarkable heterogeneity. Heterogeneity can be observed within the tumor itself, whereby multiple genetically distinct subclones can co-exist. We have shown that such differences in genetics translate into differences at the cell biological level as well (epigenome, transcriptome, proteome, metabolome and drug sensitivities) (de Boer et al. 2018) (Erdem et al. 2022). More recently, we began to dissect the heterogeneity within the tumor immune microenvironment. For instance, we identified a subgroup of AML patients that harbor a large population of tumor supportive M2 polarized macrophages in the bone marrow and these patients have the poorest prognosis. Reversely, patients with large populations of tumor suppressive M1 polarized macrophages display a much better prognosis. We also uncovered that patients with high levels of CD4+ T cells display a relatively favorable outcome.
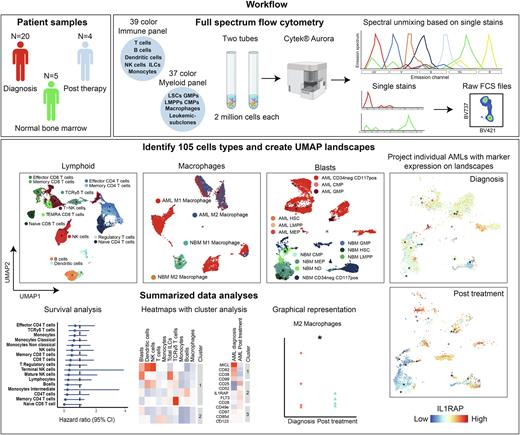
Little is known about clonal dynamics and microenvironmental changes over time or as a consequence of treatment. With the rise of new mutation-specific treatments targeting specific genetic subclones, it is important to examine the clonal composition of residual cells, especially at Measurable Residual Disease (MRD) timepoints. This can pose challenges as patients in morphologic complete remission have relatively low blast counts and limited material is available. Full spectrum flow cytometry is a novel technique that allows the use of 40 fluorescent markers in a single panel, enabling extensive immunophenotyping requiring relatively low cell numbers. We set up two antibody panels, one for immunophenotyping based on the OMIP-069 panel (39 colors) (Park et al. 2020) and one for the detection of subclones and macrophages (37 colors).
We ran both panels on AML bone marrow samples at diagnosis (n=20), after initial induction therapy (n=4) and on normal bone marrow (NBM) (n=5). Full spectrum flow cytometry showed similar results to conventional flow cytometry, although with a higher resolution. With both panels combined we could identify and quantify 105 distinct cell types. UMAP landscapes were created as a template by merging AML samples and NBM samples, on top of which activation markers and aberrant AML-specific plasma membrane (PM) protein expression profiles could be projected. Additionally, density plots of individual patient samples were generated to compare longitudinal alterations in clonal dynamics and the tumor immune microenvironment before and after treatment. Additional univariate and multivariate survival analyses were performed to link observed changes to disease progression.
At diagnosis, considerable heterogeneity amongst individual patients was observed, as expected. Patients either had a tumor supportive M2 macrophage phenotype or a tumor suppressive M1 macrophage phenotype. The lymphoid compartment was overall strongly reduced in AML compared to NBM, but was highly heterogeneous between individual patients whereby the strongest heterogeneity was noted in CD4+ and TCR gamma delta (TCRγδ) T cell subsets. Low levels of either CD4+ T cells and TCRγδ showed trends towards poorer prognosis. After induction therapy, the immune landscapes recovered and started to resemble those of NBM, most notably for T cell subsets but less so for B cells that took longer to recover. AMLs with an M2 phenotype at diagnosis shifted towards an M1 phenotype and non-classical monocytes shifted to more classical monocytes. In general, AML-specific PM markers were lost upon treatment reaching complete remission, although in some cases markers remained detectable within CD45+ blast populations. One patient containing NPMcyt (VAF=0.4) and FLT3-ITD (VAF=0.04) mutations was treated with gilteritinib in addition to cytarabine, reached morphologic complete remission whereby the FLT3-ITD mutation was lost, and the immune landscape restored. However, NPM1 droplet PCR analyses revealed the presence of residual mutant cells, which corresponded with residual cell numbers expressing aberrant CKIT/IL1RAP expression. Whether these cells will give rise to relapsed disease is currently under close investigation.
We conclude that full spectrum flow cytometry is a powerful approach that allows extensive longitudinal immunophenotyping and subclone detection requiring low cell numbers.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal